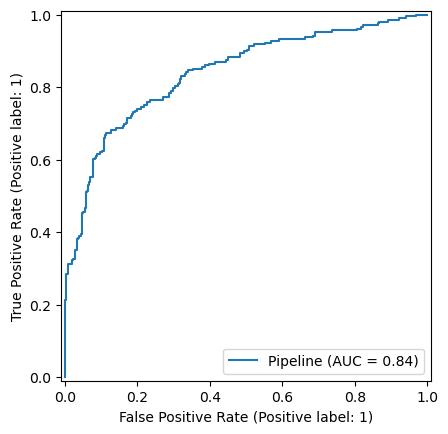
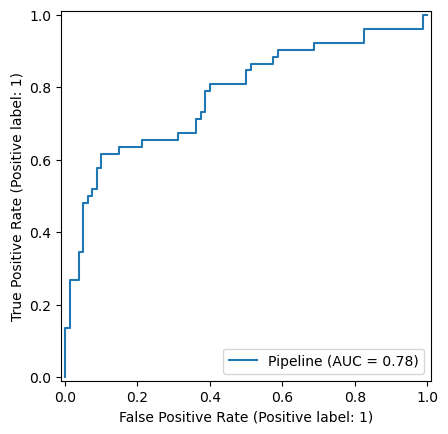
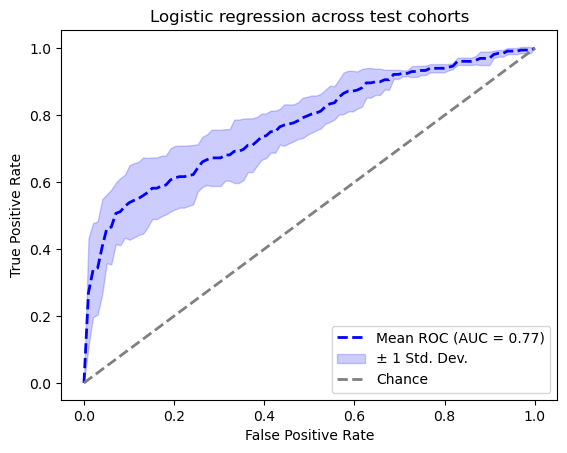
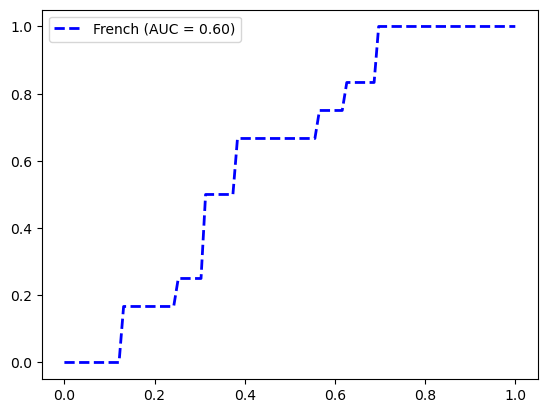
FeatureUnion(transformer_list=[('abun_part',
Pipeline(steps=[('abundance',
ColumnSelector(variables=['OTU_0',
'OTU_1',
'OTU_2',
'OTU_3',
'OTU_4',
'OTU_5',
'OTU_6',
'OTU_7',
'OTU_8',
'OTU_9',
'OTU_10',
'OTU_11',
'OTU_12',
'OTU_13',
'OTU_14',
'OTU_15',
'OTU_16',
'OTU_17',
'OTU_18',
'OTU_19',
'OTU_20',
'OTU_21',
'OTU_22',
'OTU_23',
'OTU_24',
'OTU_25',
'OTU_26',
'OTU_27',
'OTU_28',...
('pca_part',
Pipeline(steps=[('abundance',
ColumnSelector(variables=['OTU_0',
'OTU_1',
'OTU_2',
'OTU_3',
'OTU_4',
'OTU_5',
'OTU_6',
'OTU_7',
'OTU_8',
'OTU_9',
'OTU_10',
'OTU_11',
'OTU_12',
'OTU_13',
'OTU_14',
'OTU_15',
'OTU_16',
'OTU_17',
'OTU_18',
'OTU_19',
'OTU_20',
'OTU_21',
'OTU_22',
'OTU_23',
'OTU_24',
'OTU_25',
'OTU_26',
'OTU_27',
'OTU_28',
'OTU_29', ...])),
('pca', PCA())]))])